EDITORIAL
Vol. 133 No. 1510 |
SARS-CoV-2: a novel deadly virus in a globalised world
When the first reports of a cluster of mysterious atypical pneumonia cases connected to a seafood and live-animal market in Wuhan, capital of Hubei province of China appeared on 31 December 2019, visions of the 2002/2003 SARS epidemic immediately came to mind.
Full article available to subscribers
When the first reports of a cluster of mysterious atypical pneumonia cases connected to a seafood and live-animal market in Wuhan, capital of Hubei province of China appeared on 31 December 2019, visions of the 2002/2003 SARS epidemic immediately came to mind. All diagnostic tests for common respiratory pathogens, including SARS and the closely related MERS coronaviruses were negative and rumours spread that a novel coronavirus might be the cause. Closure of the market in Wuhan was initiated by Chinese officials in an attempt to contain the outbreak.
One week later, on 7 January, Chinese scientists presented data on the identification of a novel coronavirus after obtaining a whole genome sequence of the virus from a patient sample via deep metagenome sequencing.1 The virus was provisionally named 2019-nCoV (2019 novel coronavirus) and is now officially designated as SARS-CoV-2 (Severe Acute Respiratory Syndrome coronavirus 2).2 The genome sequence was shared with the scientific community on 10 January, allowing Chinese and international scientists to quickly develop real-time PCR-based detection assays. The first publicly available real-time PCR assay was published on 17 January by a German research group.3 A fervid hunt for the source of the outbreak started, with the main focus on the live animal market. It was believed that the initial transmission of the virus occurred from a wild animal to humans (zoonotic transmission) and was the cause of these pneumonia cases. However, it was believed that no human-to-human transmission had occurred. Retrospective analysis showed that almost half of the initial 41 cases had no epidemiological link to the live animal market, indicating that the live animal market might have been an accelerator of the outbreak, rather than the source. One-third of the hospitalised cases, mainly those with underlying medical conditions, had to be admitted to ICU, 12 developed acute respiratory distress syndrome similar to SARS and MERS, and six later died.4 Soon after on 18 January, the number of confirmed cases and fatalities increased and family clusters5 and infections among healthcare workers indicated that person-to-person transmission must have occurred. This led Chinese officials to introduce extensive quarantine measures and to close down metropolitan regions inhabited by tens of millions of people starting from 23 January, halting public transport, travel and trade in an attempt to control further spread, coinciding with the Chinese Lunar New Year’s festival. However, the number of reported cases grew further to almost 10,000 within four weeks (end of January) with more than 200 fatalities, suggesting a case fatality rate of approximately 2%, which is low compared to 10% for SARS-CoV and up to 35% for MERS-CoV. In comparison, 298,120 laboratory-confirmed cases and 812 deaths were reported in Australia during the 2019 influenza season, with a case fatality rate of 0.27%.
The risk of mortality due to a pathogen is heavily influenced by the characteristics of the affected population, including age and available healthcare resources. Measles illustrates this pointedly as the risk of fatality due to the virus in developed countries increases 100-fold in developing countries, influenced by limited healthcare resources. The same applies to the basic reproduction number R0 (R nought) that describes the expected number of cases directly generated by one infected person in a susceptible population. For SARS-CoV-2, R0 has been estimated to be 2.68,6 but this number is not constant and is affected by infection control practices such as quarantine and respiratory hygiene, which can decrease R0 and stop an outbreak. In a population without these countermeasures R0 can be much higher.
Figure 1: Phylogenetic tree of coronaviruses including novel coronavirus SARS-CoV-2 (solid box outline) and common cold coronaviruses OC43, HKU1, 229E and NL63 (dashed box outline).
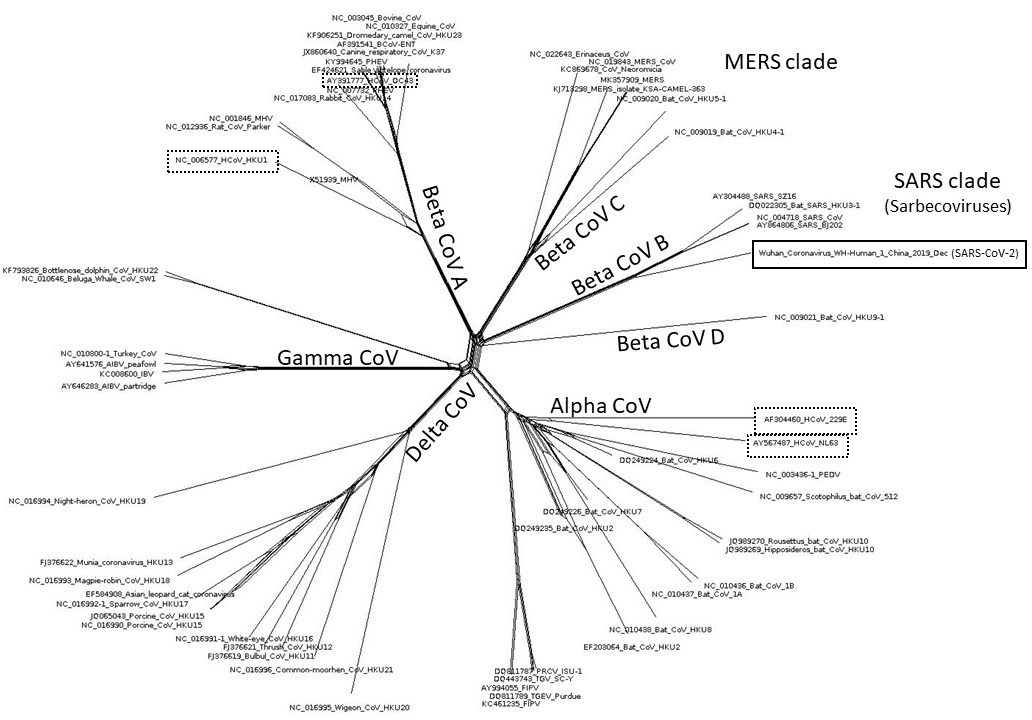
In China, the country’s impressive rapid response and quarantine measures have not stopped the virus spreading widely to all Chinese provinces, crossing borders to neighbouring countries and via international travel to distant continents. After initial hesitancy the WHO finally declared the outbreak a public health emergency of international concern (PHEIC) on 30 of January.7 Human-to-human-transmission outside of China had been detected, while spread to countries with limited healthcare resources was perceived as a significant additional challenge.
SARS-CoV-2
Coronaviruses are a big family of RNA viruses with a broad host spectrum including birds, sea mammals, rodents, civet cats, raccoon dogs, camels, bats, reptiles, fish and humans. So far coronaviruses pathogenic to humans include four viruses with global distribution, OC43, HKU1, NL63 and 229E contributing to up to 30% of upper respiratory tract infections (URTI) each year similar to the common cold and two additional localised/sporadic coronaviruses, SARS-CoV and MERS-CoV, that infect the lower respiratory tract and can cause acute respiratory distress syndrome (ARDS). Phylogenetic analysis based on a sequence alignment of whole genome sequences revealed that SARS-CoV-2 is a member of the SARS clade (lineage B) of beta coronaviruses (Sarbecoviruses) with 79.5% sequence similarity to human SARS virus and 96.2% sequence similarity to a bat coronavirus isolated from horseshoe bats in the Yunnan province of China.8 This suggests that bats, the known source for SARS-CoV might also be the zoonotic reservoir of SARS-CoV-2. It is highly likely that an intermediate host animal, possibly a pangolin (so far unpublished) traded at the Wuhan live-animal market, has transmitted the virus from bats into an immunological naïve human population. Metagenomic surveys show that bats are among the most abundant sources for novel viral sequences and that more than 200 strains of coronaviruses have been described from bats,9,10 50 of which are SARS related. Bats are also the reservoir host for many highly pathogenic viruses like Ebola, Marburg, Nipah, Hendra and Rabies. They are one of the oldest mammalian groups with a unique immune system, which allows them to be asymptomatic carriers of these viruses, with the exception of Rabies. In addition, they have a long lifespan, often live in large colonies and can fly across large geographical regions. Like Ebola virus, where outbreaks frequently originate from bush meat markets in Africa, this coronavirus outbreak seems to be linked to live-animal markets in China. The Wildlife Conservation Society recently emphasised that poorly regulated live-animal markets mixed with illegal wildlife trade offer a unique opportunity for viruses to spill over from wildlife hosts into the human population.11 But in contrast to most African countries, China is well connected via international flights with the rest of the world. The modern phenomenon of globalisation has the potential to assist the efficient and rapid dispersion of this novel pathogen.
Bats and humans share the same cellular receptor, ACE2, which interacts with the SARS-CoV and SARS-related coronavirus spike protein, enabling entry into and efficient replication in primary human airway cells deep in the lungs and cells of the intestine. Preliminary data suggest that SARS-CoV-2 is using the same cellular receptor for cell entry.1,12,13 In addition to coughing and sneezing, transmission via the faecal-oral route may also be possible. But there appear to be additional host factors that determine the severity of the disease, since most infected people with SARS-CoV, MERS-CoV or SARS-CoV-2 only display mild symptoms and mainly people with comorbidities develop more severe clinical outcomes.
Interestingly, coronaviruses have the biggest genomes of all RNA viruses, with genome sizes up to 32 kilobases (kb), coding for a large number of proteins including 4 structural, 16 non-structural and 6–8 accessory proteins, some of which are counteracting the innate immune response. It has been shown that the ORF3b-gene of SARS-CoV-2 encodes a completely novel putative protein of unknown function.14 Usually, the larger the size of an RNA genome, the bigger the impacts on the fitness of the virus due to the high mutation rate of RNA viruses and accumulation of unfavourable mutations. Coronaviruses have adapted by encoding an exonuclease conferring proof-reading activity and by using mechanisms like recombination (possibly even with members of different virus families),15 horizontal gene transfer, gene duplication and alternative open reading frames to expand their genetic variability and capacity to infect new hosts.16 What we have seen so far indicates that SARS-CoV-2 has maintained viral fitness and is readily transmitted from human to human.
Coronavirus disease (COVID-19)
Knowledge of COVID-19, the disease caused by SARS-CoV-2, its severity and the range of symptoms is constantly evolving.
Reported clinical findings include fever (less frequent than in SARS and MERS) and respiratory symptoms, most prominently a dry cough. Other findings include ground-glass radiological lung opacities, normal or reduced leukocyte and thrombocyte counts, hypoxaemia, deranged liver and renal function.4 In addition, SARS-CoV-2 causes severe respiratory illness in approximately 16–20% of all infected cases, a much higher rate than influenza. Even in developed countries, high numbers of patients with severe respiratory illness can overwhelm the healthcare systems. In a case report investigating a familial cluster of COVID-19 pneumonia, some members of the family also presented with diarrhoea.5 It is likely that SARS-CoV-2 infections, like other respiratory pathogens, will range from asymptomatic infection to severe acute respiratory syndrome, potentially associated with more severe outcomes in patients with existing comorbidities. There is currently little or no information on infections in pregnant women.
Because of the non-specific nature of these symptoms, it is essential that healthcare practitioners obtain a detailed travel history of suspected cases with respiratory symptoms to ensure that those meeting the current suspected case definition undergo testing for a range of viral respiratory pathogens, including SARS-CoV-2.17 The specimen types currently recommended for testing are nasopharyngeal and oropharyngeal swabs in ambulatory patients and sputum or endotracheal aspirate in patients with more severe respiratory disease.18 It is important to note that at this point in time there is no robust evidence about the optimal specimen for testing. Most respiratory viral pathogens replicate in the nasopharyngeal epithelium. It was observed during the pandemic with Influenza A(H1N1) 2009 that in severe lower respiratory disease nasopharyngeal swabs could give false-negative results.19 It remains to be seen to what extent this applies also to SARS-CoV-2.
Not enough is known about the usefulness of non-respiratory specimens including serum or plasma, urine or faecal specimens for the diagnosis of COVID-19 but guidance will be updated as new information comes to light.
Our understanding of the transmission of this virus and its virulence is evolving at a fast pace. The incubation period range for SARS-CoV-2 infection is between two days and 12 days, and in some conservative models up to 16 days, with a mean incubation time of 6.4 days.20 Another study describes an incubation period range of 0–24 days with a mean incubation time of three days.21 While it appeared that droplet spread was the transmission route, new information suggests aerosol transmission is occurring. In addition, in contrast to other enveloped viruses, coronaviruses are fairly stable for several days on surfaces such as door handles,22 probably the result of the high amount of spike proteins in their envelope. During the SARS and MERS outbreaks, ‘super-spreaders’ caused significant nosocomial outbreaks, with up to 82 cases occurring in a single hospital related to one infected person.23 In an investigation of hospitalised COVID-19 patients from Wuhan, 57 out of 138 (41%) patients were associated with presumed nosocomial transmission.24 This reinforces the importance of adherence to strict infection control practices in the hospital setting.
However, the limited understanding of the transmission dynamics of SARS-CoV-2 has made the development of robust and meaningful guidelines for infection control, quarantine and containment very challenging.
Where to from here?
The emergence of SARS-CoV-2 is a new reminder of the importance of monitoring the unintended consequences of globalisation of the world we live in. As the ability to fly across the globe is an expectation for many of us, it also harbours challenges, such as containment of potential pathogens. It took the Plague years to spread across Europe in the 14th and 15th century, while it took SARS-CoV-2 only 13 days from the identification of a cluster of cases in Wuhan to spread to countries outside China. As of 12 February 2020, 44,685 laboratory-confirmed cases have been reported from mainland China and 518 confirmed cases in 27 countries outside of China with 1,116 fatalities. The actual number of infected cases is likely to be much higher.
In line with the International Health Regulations (2005) facilitating the timely sharing of information with the World Health Organization (WHO),7 the international scientific community and the world, the concept of ‘One Health’ encourages us to look at the interaction of our environment, animal and human health. The example of COVID-19 illustrates well that emerging infectious diseases are not the sole responsibility of the medical profession. It requires rapid and robust communication systems across countries, across political systems and across professional boundaries to ensure that novel pathogens are quickly identified, characterised and contained, if possible.
Authors
Meik Dilcher, Microbiology Department, Canterbury Health Laboratories, Christchurch; Anja Werno, Microbiology Department, Canterbury Health Laboratories, Christchurch; Lance C Jennings, Department of Pathology and Biomedical Science, University of Otago, Christchurch.Correspondence
Meik Dilcher, Microbiology Department, Canterbury Health Laboratories, 525 Hagley Avenue, Christchurch 8011.Correspondence email
meik.dilcher@cdhb.health.nzCompeting interests
Nil.1. Peng Zhou, X-LY, Xian-Guang Wang, Ben Hu, Lei Zhang, Wei Zhang, Hao-Rui Si, Yan Zhu Bei Li, Chao-Lin Huang, Hui-Dong Chen, Jing Chen, Yun Luo, Hua Guo, Ren-Di Jiang, Mei-Qin Liu, Ying Chen, Xu-Rui Shen, Xi Wang, Xiao-Shuang Zheng, Kai Zhao, Quan-Jiao Chen, Fei Deng, Lin-Lin Liu, Bing Yan, Fa-Xian Zhan, Yan-Yi Wang, Geng-Fu Xiao, Zheng-Li Shi, Discovery of a novel coronavirus associated with the recent pneumonia outbreak in humans and its potential bat origin. bioRxiv, 2020. http://dx.doi.org/10.1101/2020.01.22.9149522.
2. Gorbalenya A. Severe acute respiratory syndrome-related coronavirus - The species and its viruses, a statement of the Coronavirus Study Group. bioRxiv, 2020. https://doi.org/10.1101/2020.02.07.9378623.
3. Corman VM, et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill, 2020; 25(3).
4. Huang C, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet, 2020.
5. Chan JF, et al. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet, 2020.
6. Wu JT, Leung K, Leung GM, Nowcasting and forecasting the potential domestic and international spread of the 2019-nCoV outbreak originating in Wuhan, China: a modelling study. Lancet, 2020.
7. WHO, Statement on the second meeting of the International Health Regulations (2005) Emergency Committee regarding the outbreak of novel coronavirus (2019-nCoV). http://www.who.int/news-room/detail/30-01-2020-statement-on-the-second-meeting-of-the-international-health-regulations-(2005)-emergency-committee-regarding-the-outbreak-of-novel-coronavirus-(2019-ncov), 2020.
8. Peng Zhou, X-LY, Xian-Guang Wang, Ben Hu, Lei Zhang, Wei Zhang, Hao-Rui Si, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature, 2020.
9. Hu B, et al. Discovery of a rich gene pool of bat SARS-related coronaviruses provides new insights into the origin of SARS coronavirus. PLoS Pathog, 2017; 13(11): p. e1006698.
10. Banerjee A, et al. Bats and Coronaviruses. Viruses, 2019; 11(1).
11. Walzer C. Statement of the Wildlife Conservation Society on the Wuhan coronavirus. http://www.wcs.org/get-involved/updates/a-primer-on-the-coronavirus, 2020.
12. Zhou P, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature, 2020.
13. Lu R, et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet, 2020.
14. Jasper Fuk-Woo Chan, K-HK, Zheng Zhu, Hin Chu, Kelvin Kai-Wang To, Shuofeng Yuan, Kwok-Yung Yuen, Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerging Microbes & Infection, 2020; 9(1): p. 221–236.
15. Huang C, et al. A Bat-Derived Putative Cross-Family Recombinant Coronavirus with a Reovirus Gene. PLoS Pathog, 2016; 12(9): p. e1005883.
16. Menachery VD, Graham RL, Baric RS. Jumping species-a mechanism for coronavirus persistence and survival. Curr Opin Virol, 2017; 23: p. 1–7.
17. Ministry of Health, NZ, Case definition of 2019-nCoV infection. http://www.health.govt.nz/our-work/diseases-and-conditions/novel-coronavirus-covid-19/case-definition-2019-ncov-infection 2020.
18. WHO, Laboratory testing of human suspected cases of novel coronavirus (nCoV) infection. Interim Guidance, 2020. WHO/2019-nCoV/laboratory/2020.1.
19. Mulrennan S, et al. Pandemic influenza (H1N1) 2009 pneumonia: CURB-65 score for predicting severity and nasopharyngeal sampling for diagnosis are unreliable. PLoS One, 2010; 5(9): p. e12849.
20. Backer JK, D; Wallinga J. Incubation period of 2019 novel coronavirus (2019-nCoV) infections among travellers from Wuhan, China, 20-28 January 2020. Euro Surveill, 2020; 25(5).
21. Wei-jie Guan, Z-yN, Yu Hu, Wen-hua Liang, Chun-quan Ou, Jian-xing He, Lei Liu, Hong Shan, Chun-liang Lei, David SC Hui, Bin Du, Lan-juan Li, Guang Zeng, Kowk-Yung Yuen, Ru-chong Chen, Chun-li Tang, Tao Wang, Ping-yan Chen, Jie Xiang, Shi-yue Li, Jin-lin Wang, Zi-jing Liang, Yi-xiang Peng, Li Wei, Yong Liu, Ya-hua Hu, Peng Peng, Jian-ming Wang, Ji-yang Liu, Zhong Chen, Gang Li, Zhi-jian Zheng, Shao-qin Qiu, Jie Luo, Chang-jiang Ye, Shao-yong Zhu, Nan-shan Zhong. Clinical characteristics of 2019 novel coronavirus infection in China. medRxiv, 2020. http://doi.org/10.1101/2020.02.06.20020974
22. GT, D; Pfaender S, Steinmann E. Persistence of coronaviruses on inanimate surfaces and its inactivation with biocidal agents. The Journal of Hospital Infection, 2020.
23. Cho SY, et al. MERS-CoV outbreak following a single patient exposure in an emergency room in South Korea: an epidemiological outbreak study. Lancet, 2016; 388(10048): p. 994–1001.
24. Wang D, et al. Clinical Characteristics of 138 Hospitalized Patients With 2019 Novel Coronavirus-Infected Pneumonia in Wuhan, China. JAMA, 2020.
